Scientists have successfully and repeatedly encoded, stored and erased digital data within the DNA of living cells.
The Stanford team reapplied natural enzymes adapted from bacteria to flip specific sequences of DNA back and forth at will to create what’s essentially a bit: if the DNA section points in one direction, it’s a zero, while if it points the other way, it’s a one.
“It took us three years and 750 tries to make it work, but we finally did it,” says Dr Jerome Bonnet.
The team suggests that the technique could be a powerful tool for studying cancer, aging, organismal development and even the natural environment. Researchers could count how many times a cell divides, for instance, perhaps giving the ability to turn off cells before they become cancerous.
But it could also form the basis of as non-volatile memory – data storage that can retain information without consuming power – which is referred to in biotechnology as recombinase-mediated DNA inversion.
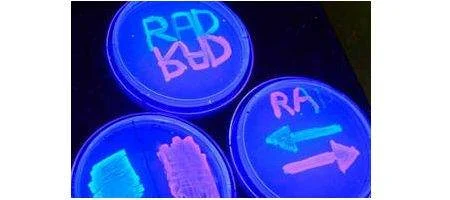
The team calls its device a ‘recombinase addressable data’ module, or RAD. They used it to modify a particular section of DNA with microbes to determine how the one-celled organisms will fluoresce under ultraviolet light. The microbes glow red or green depending upon the orientation of the section of DNA – and, using RAD, the engineers can flip the section back and forth at will.
The team has now tested RAD modules in single microbes that have doubled more than 100 times – and the switch has held. They’ve also switched the latch and watched a cell double 90 times, and set it back. The latch will even store information when the enzymes are not present. In short, they say, RAD works – it’s reliable and rewritable.
The team’s now hoping to move on from the current single bit to a full byte of programmable genetic data storage – although this won’t be easy.
“Such systems will likely be 10 to 50 times more complicated than current state-of-the-art genetic engineering projects,” says assistant professor Dr Drew Endy.
“We’re probably looking at a decade from when we started to get to a full byte. But by focusing today on tools that improve the engineering cycle at the heart of biotechnology, we’ll help make all future engineering of biology easier, and that will lead us to much more interesting places.”






